The prolonged misuse of antibiotics has led to the continuous emergence of drug-resistant bacteria, making it urgent to identify new antimicrobial targets and develop novel antibiotics. Bacterial division is a vital physiological process for bacterial survival, and the proteins involved have always been crucial targets for new antibiotics. The core process of bacterial division involves the synthesis of the cell wall peptidoglycan layer, mediated by the peptidoglycan glycosyltransferase FtsW and the monofunctional transpeptidase FtsI. FtsW polymerizes the peptidoglycan precursor Lipid II into chains, and FtsI cross-links these chains to form a new peptidoglycan layer. The FtsQLB complex also plays an important regulatory role in this process and can form the supercomplex FtsWIQLB with FtsWI. Studies show that the functional loss of FtsWI or FtsQLB leads to bacterial death. However, the molecular mechanisms by which FtsWI synthesizes peptidoglycan and FtsQLB regulates this process are not yet clear. Therefore, research on the structure and mechanisms of the FtsWIQLB complex will greatly promote the development of antibiotics targeting this complex.
This study utilized single-particle cryo-electron microscopy to successfully resolve the high-resolution structure of the bacterial division complex FtsWIQLB from Pseudomonas aeruginosa and conducted a series of functional analyses. Structural analysis revealed that the ECL4 of FtsW plays a significant functional role. Additionally, FtsW possesses a distinct hydrophobic cavity, and sequence analysis has shown that this cavity contains important conserved amino acids. Therefore, this study successfully obtained ftsW gene knockout strains using  λ−Red homologous recombination and CRISPR-Cas9 technology, and conducted mutations on the conserved amino acids in the cavity of FtsW. Through bacterial plate survival experiments and morphological observation experiments, we identified critical functional sites of FtsW and preliminarily elucidated the binding mechanism of FtsW with its substrate, Lipid II.
λ−Red homologous recombination and CRISPR-Cas9 technology, and conducted mutations on the conserved amino acids in the cavity of FtsW. Through bacterial plate survival experiments and morphological observation experiments, we identified critical functional sites of FtsW and preliminarily elucidated the binding mechanism of FtsW with its substrate, Lipid II.
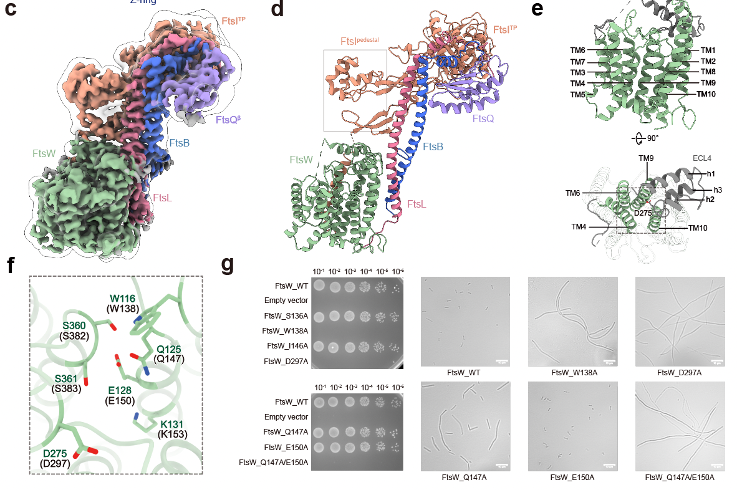
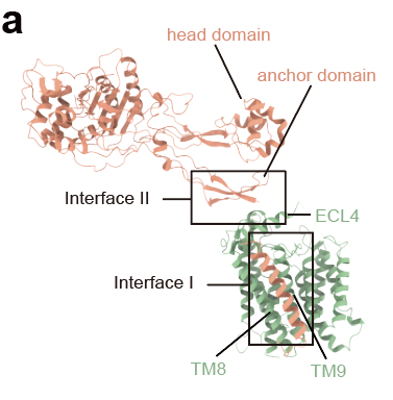
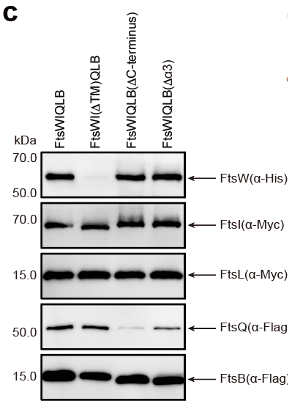
Additionally, based on the structural information, FtsW and FtsI have two important interaction interfaces: one located in the transmembrane region and the other near the periplasmic membrane area. The C-terminus of FtsQLB has significant interactions with FtsI. Through in vitro complex stability experiments, it was found that the single transmembrane helix of FtsI and the C-terminal region of FtsB are crucial for the stability of this complex.
Bacterial division is an important process for maintaining normal physiological activities in bacteria, and studying the structure and mechanisms of related proteins can lay an important foundation for the development of new antibiotics.
This research achievement was successfully published in Cell Discovery on January 4, 2024. Doctoral students Yang Lili and Chen Yujiao from the National Key Laboratory of Biotherapy (Sichuan University), along with Dr. Chang Shenghai, an engineer at the Cryo-Electron Microscopy Center of Zhejiang University, are co-first authors of this paper.