
Recently, the research team of Haohao Dong, State Key Laboratory of Biotherapy of West China Hospital of our university, published a research paper entitled "Structural basis for bacterial lipoprotein relocation by the transporter LolCDE" in the international journal Nature Structural & Molecular Biology. The research work was completed by the team of researcher Haohao Dong, the team of Professor Xing Zhang, director of the Cryo-EM Center of Zhejiang University School of Medicine, and the team of Professor Changjiang Dong of the University of East Anglia School of Medicine. Dr. Xiaodi Tang, associate professor of the State Key Laboratory of Biotherapy of our university, Dr. Shenghai Chang, engineer of the cryo-EM center of Zhejiang University, Ke Zhang, laboratory assistant of our university, and Qinghua Luo, a doctoral student, were the first authors. The study was strongly supported by Prof. Xiawei Wei and Prof. Xiaofeng Zhu from Sichuan University and Prof. Zhengyu Zhang from Wuhan University. The State Key Laboratory of Biotherapy/National Clinical Research Center for Geriatric Diseases is the first author and corresponding author of the paper.
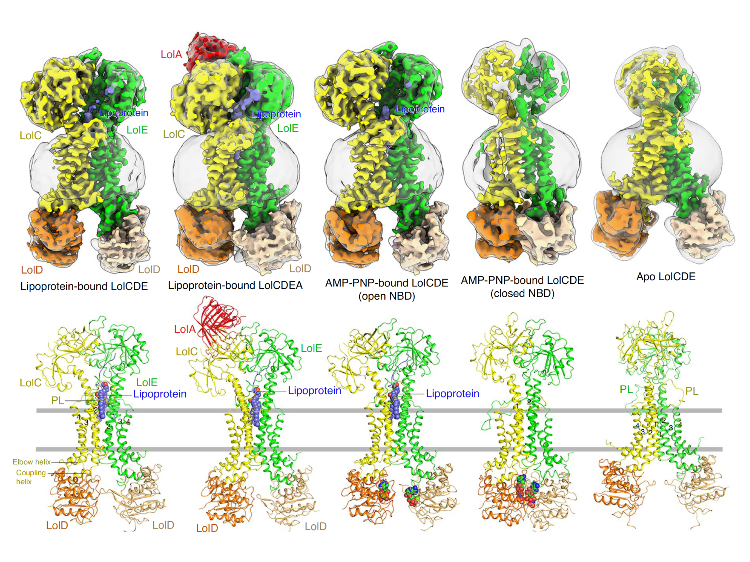
In this study, LolCDE (apo), a protein machine used by multidrug-resistant bacteria-gram-negative bacteria to transport the lipoprotein components of the outer membrane, and six high-resolution cryo-EM structures combined with substrate lipoprotein, ADP, ATP isomeric inhibitor AMP-PNP and periplasmic protein LolA, were analyzed, demonstrating four intermediate conformations in their transport functions. At the same time, the study further revealed the functional sites of identifying and transporting lipoproteins through biochemical function experiments, which provided an important scientific basis for the design of new antibacterial drugs targeting this protein machine.
Gram-negative bacteria represented by Pseudomonas aeruginosa, Enterobacter and Acinetobacter baumannii are clinically common, and they are also a class of pathogenic bacteria with the highest concentration of drug-resistant bacteria, and their unique outer membrane structure is the most important protective barrier to prevent antibiotics from entering cells. Lipoproteins located in the outer membrane of gram-negative bacteria are important, which participate in the transport and assembly of many outer membrane components (including lipopolysaccharides, phospholipids, outer membrane β-barrel proteins and lipoproteins, etc.) to maintain the asymmetry, integrity and physiological functions of the lipid bilayer of the bacterial outer membrane, which is the core of the entire outer membrane formation and plays an important role in the growth and survival of drug-resistant bacteria. LolCDEAB is responsible for transporting dozens of outer membrane lipoproteins from the bacterial inner membrane into the outer membrane. Mislocalization of outer membrane lipoproteins in the inner cell membrane can lead to bacterial death, so understanding the localization mechanism of outer membrane lipoproteins and discovering their drug targets for positioning protein machinery is of great significance for the development of potential new antibiotics.
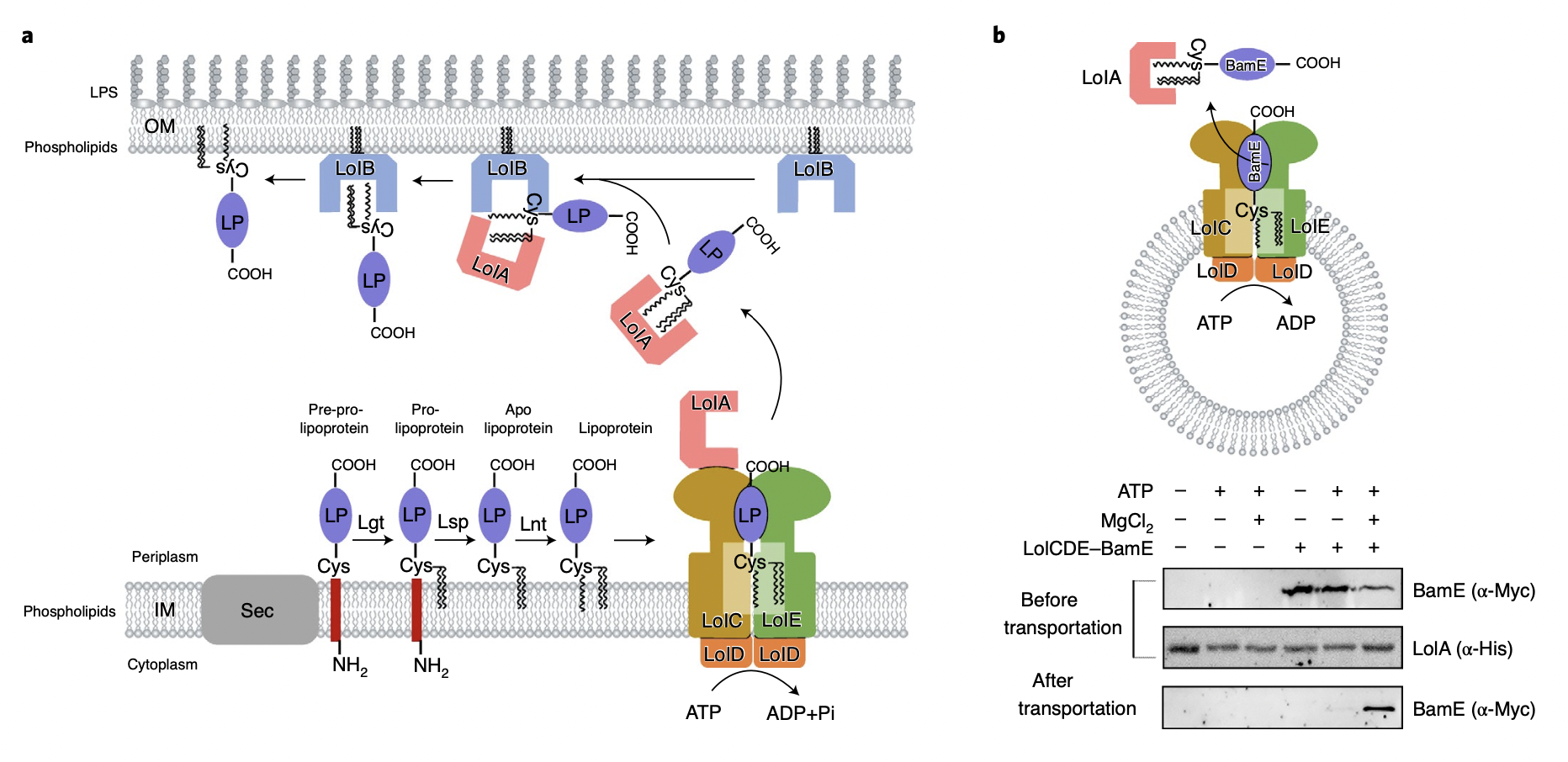
The lipoproteins of gram-negative bacteria (including endometrial lipoprotein and outer membrane lipoprotein) are on the periplasmic side of the bacterial inner membrane after intracellular synthesis of bacteria, and the protein machine LolCDE classifies whether it is an outer membrane lipoprotein by recognizing the transport signal of the N-terminus of lipoprotein, thereby extracting the target substrate from the inner membrane and transporting it to the transport protein LolA in the periplasm. The research team used single-particle cryo-EM technology to analyze the molecular structure of LolCDE responsible for this step for the first time, revealing the molecular details and interaction relationships between the functional domains of the protein complex and the ligand. By capturing different functional conformations of LolCDE in different transport stages, including apo-state, lipoprotein-binding state, ATP-bound dimerized state, and LolA-bound pre-release state, a complete set of mechanistic changes in lipoprotein transport is demonstrated.
The findings of this study confirm that LolCDE differs from the symmetric allosteric mechanism of other bacterial ABC transporters, and its transmembrane subunits LolC and LolE have different functions in participating in lipoprotein transport. LolE is responsible for identifying and locating lipoproteins, while LolC acts as a mechanical lever to conduct conformational changes from NBD (nucleotide-binding domains) to PD (periplasmic domains) before and after ATP binding and hydrolysis, by coordinating the various functional domains of the protein machine for lipoprotein extraction and transport. The structural homology of LolCDE with other type VII ABC transporters suggests that this mechanotransduction mechanism may be common in such transporter families. In conclusion, through the molecular details of the binding of the protein machine LolCDE to the substrate lipoprotein, the simulated transport experiments of fixed-point single mutation and construction of protein phospholipid vesicles in vitro were carried out, and multiple functional sites that can inhibit bacterial growth by hindering their transport function were discovered, which could be used as drug targets for the development of inhibitors. This study has important implications for understanding the mechanism of action of other type VII ABC transporters and facilitating drug development for the treatment of multidrug-resistant gram-negative bacteria.